Deeplex for prediction of M. tuberculosis resistance and NTM identification
A deep sequencing-based assay for the prediction of M. tuberculosis resistance to 15 antibiotics and NTM identification
Deeplex® Myc-TB is an innovative assay for Mycobacterium tuberculosis complex resistance prediction to 15 antibiotics based on next-generation sequencing, with a turnaround time less than 48 hours, and directly applicable on clinical samples.
Our test predicts extensive drug resistance (XDR) as recently redefined by the World Health Organization, combining resistance to any fluoroquinolone, to bedaquiline and/or linezolid, in addition to resistance to rifampicin or to rifampicin and isoniazid (multidrug resistance).
The kit also enables the identification of more than 100 nontuberculous mycobacteria at (sub)species or species complex level, including most clinically relevant species such as M. kansasii, M. intracellulare and M. avium complex.
Discover the mycobacteria identified by Deeplex
List of mycobacteria included in the hsp65 database of Deeplex® Myc-TB for Mycobacterium tuberculosis complex resistance prediction. Species identification of highlighted mycobacteria was confirmed by an independent reference, as described in Dai et al., J Clin Microbiol, 2011.
M. abscessus (subsp abscessus, bolletii)
M. africanum
M. agri
M. aichiense
M. algericum
M. alvei
M. aromaticivorans
M. arosiense
M. arupense
M. asiaticum
M. aubagnense
M. aurum
M. austroafricanum
M. avium (subsp avium, paratuberculosis, silvaticum)
M. boenickei
M. bohemicum
M. botniense
M. bouchedurhonense
M. bourgelatii
M. bovis
M. bovis BCG
M. branderi
M. brisbanense
M. brumae
M. canariasense
M. canettii (CIPT140010059, STB-D, E, G, H, I, J, K, L)
M. caprae
M. celatum
M. chelonae
M. chimaera
M. chitae
M. chlorophenolicum
M. chubuense
M. colombiense
M. conceptionense
M. confluentis
M. conspicuum
M. cookii
M. cosmeticum
M. crocinum
M. diernhoferi
M. doricum
M. duvalii
M. elephantis
M. europaeum
M. fallax
M. farcinogenes
M. flavescens
M. florentinum
M. fluoranthenivorans
M. fortuitum (subsp acetamidolyticum, fortuitum)
M. fragae
M. frederiksbergense
M. gadium
M. gastri
M. genavense
M. gilvum
M. goodii
M. gordonae
M. haemophilum
M. hassiacum
M. heckeshornense
M. heidelbergense
M. hiberniae
M. hodleri
M. holsaticum
M. houstonense
M. immunogenum
M. insubricum
M. interjectum
M. intermedium
M. intracellulare
M. kansasii
M. komossense
M. kubicae
M. kumamotonense
M. kyorinense
M. lacus
M. lentiflavum
M. lepraemurium
M. leprae
M. llatzerense
M. madagascariense
M. mageritense
M. malmoense
M. mantenii
M. marinum
M. marinum M
M. marseillense
M. massiliense
M. microti
M. monacense
M. montefiorense
M. moriokaense
M. mucogenicum
M. murale
M. nebraskense
M. neoaurum
M. neworleansense
M. onchromogenicum
M. noviomagense
M. novocastrense
M. obuense
M. pallens
M. palustre
M. paraffinicum
M. parafortuitum
M. parascrofulaceum
M. paraseoulense
M. parmense
M. peregrinum
M. phlei
M. phocaicum
M. porcinum
M. poriferae
M. pseudoshottsii
M. psychrotolerans
M. pulveris
M. pyrenivorans
M. rhodesiae
M. riyadhense
M. rufum
M. rutilum
M. salmoniphilum
M. saskatchewanense
M. scrofulaceum
M. sediminis
M. senegalense
M. senuense
M. seoulense
M. septicum
M. setense
M. sherrisii
M. shimoidei
M. shinjukuense
M. shottsii
M. simiae
M. smegmatis
M. sphagni
M. stomatepiae
M. szulgai
M. terrae
M. thermoresistibile
M. timonense
M. tokaiense
M. triplex
M. triviale
M. tuberculosis complex
M. tusciae
M. ulcerans
M. vaccae
M. vanbaalenii
M. vulneris
M. wolinskyi
M. xenopi
M. yongonense
Proven high accuracy across a broad range of NTM species
Based on sequence identity of the hsp65 gene, Deeplex® Myc-TB NTM identification accuracy has been tested over 73 different NTM species in Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs, Jouet A et al., Eur Respir J, 2020, with more than 93% of strains concordantly identified at (sub)species or species complex levels by both reference and Deeplex® Myc-TB testing.
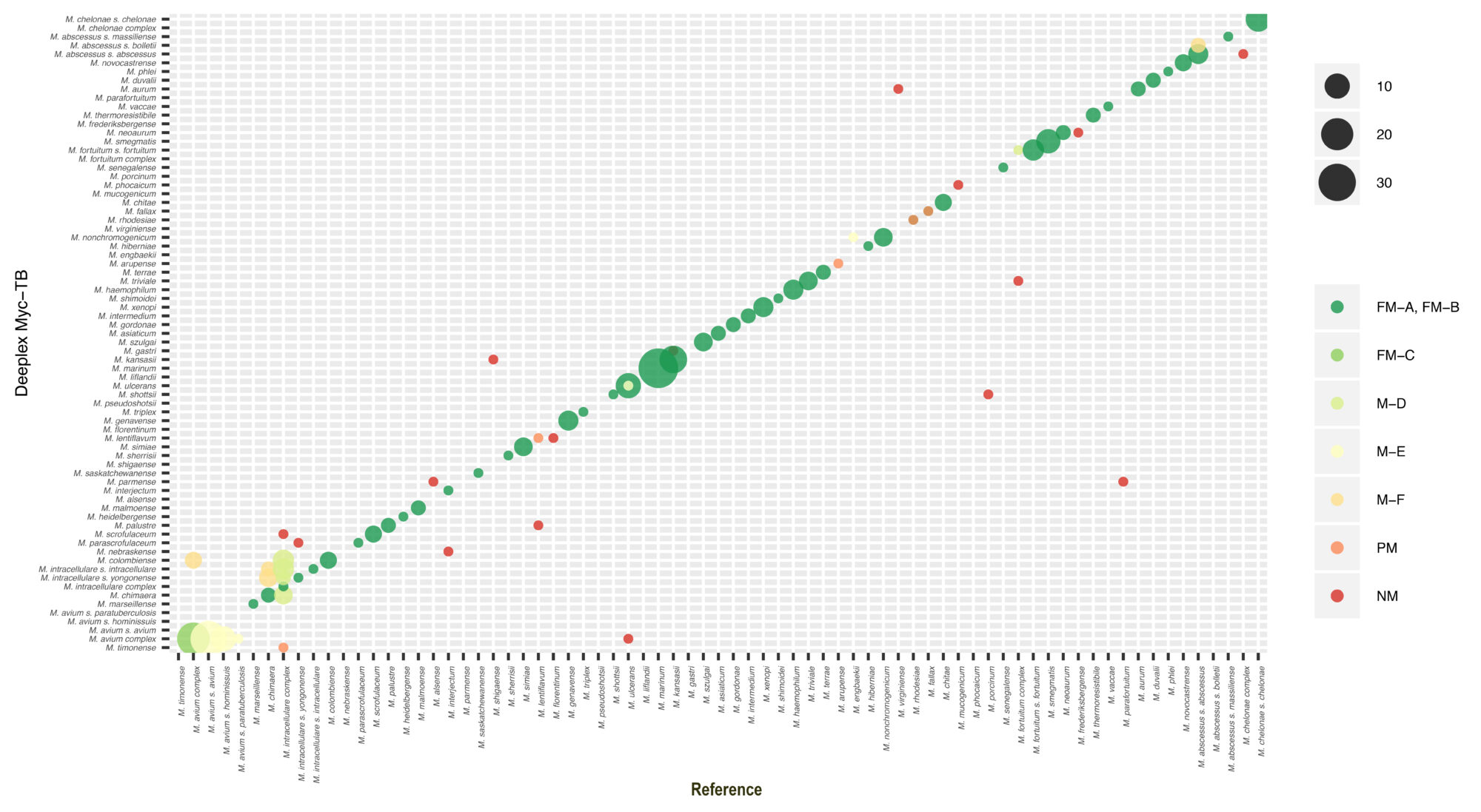
Identification of nontuberculous mycobacterial species by Deeplex® Myc-TB versus reference identification
The results were based on hsp65 best match analysis, complemented by specific SNP detection in 16S (rrs) and 23S rDNA (rrl) targets for M. kansasii and M. chelonae strains, while reference identification was based on rpoB and/or 16S rDNA Sanger sequencing results, phenotypic profiling and/or type strain status. The number of isolates studied per taxon (complex, species or subspecies) is proportional to associated circle sizes.
FM-A, FM-C: Full match at same taxonomic level (complex, species or subspecies), FM-B: Full match at (sub)species level but with a clonal mix identified by Deeplex®, MD: Match at complex level, sublevel provided by Deeplex®, M-E: Match at complex level, sublevel provided by the reference, M-F: Match at complex level, different subspecies, PM: Partial match, several possible species identified by Deeplex®, NM: No match. Non-tuberculosis mycobacterial taxa are ordered phylogenetically, according Tortoli et al. Infect Genet Evol 2019;75:103983.
The remaining 6% of the strains, which had taxonomically discordant results even at the complex level between both methods, mostly consisted of single discordant cases among otherwise partially/fully concordant strains for a species or species rarely involved in infections.
However, for some of these few individual isolates of otherwise well-identified species, the correctness of the Deeplex® Myc-TB identification was actually often possible or probable, as conflicting or ambiguous identifications were seen between the rpoB and 16S rDNA reference probes, with one or the other partially or fully matching the Deeplex® Myc-TB result. The same often held true for the residual isolates with discrepant subspecies within a matching complex.
2021
Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs
