Tackle Infectious Diseases and Antimicrobial Resistance
Infectious diseases are a major global health problem, accounting for one in five deaths worldwide and a quarter of the annual global disease burden (in terms of years of life lost due to premature mortality or physical disabilities).
Get Clinically Actionable Information from Microbial DNA
Deeplex® engineering combines deep multi-target DNA sequencing, automated bioinformatic analysis and easy reporting for microbial identification and extensive detection of drug resistance.
Powerful Solutions for Clinicians and Researchers
By overcoming major limitations of conventional phenotypic and molecular testing, Deeplex® products represent novel powerful solutions for improving the clinical management and the monitoring of drug resistant infectious diseases.
FAST.
SECURE.
EASY TO INTERPRET.
The Deeplex® engineering is a next-generation sequencing based technology, developed by GenoScreen, for accurate characterization of specific pathogens and prediction of their antimicrobial resistance.
This technology provides clinicians and researchers with extensive information to tackle major infectious diseases worldwide.
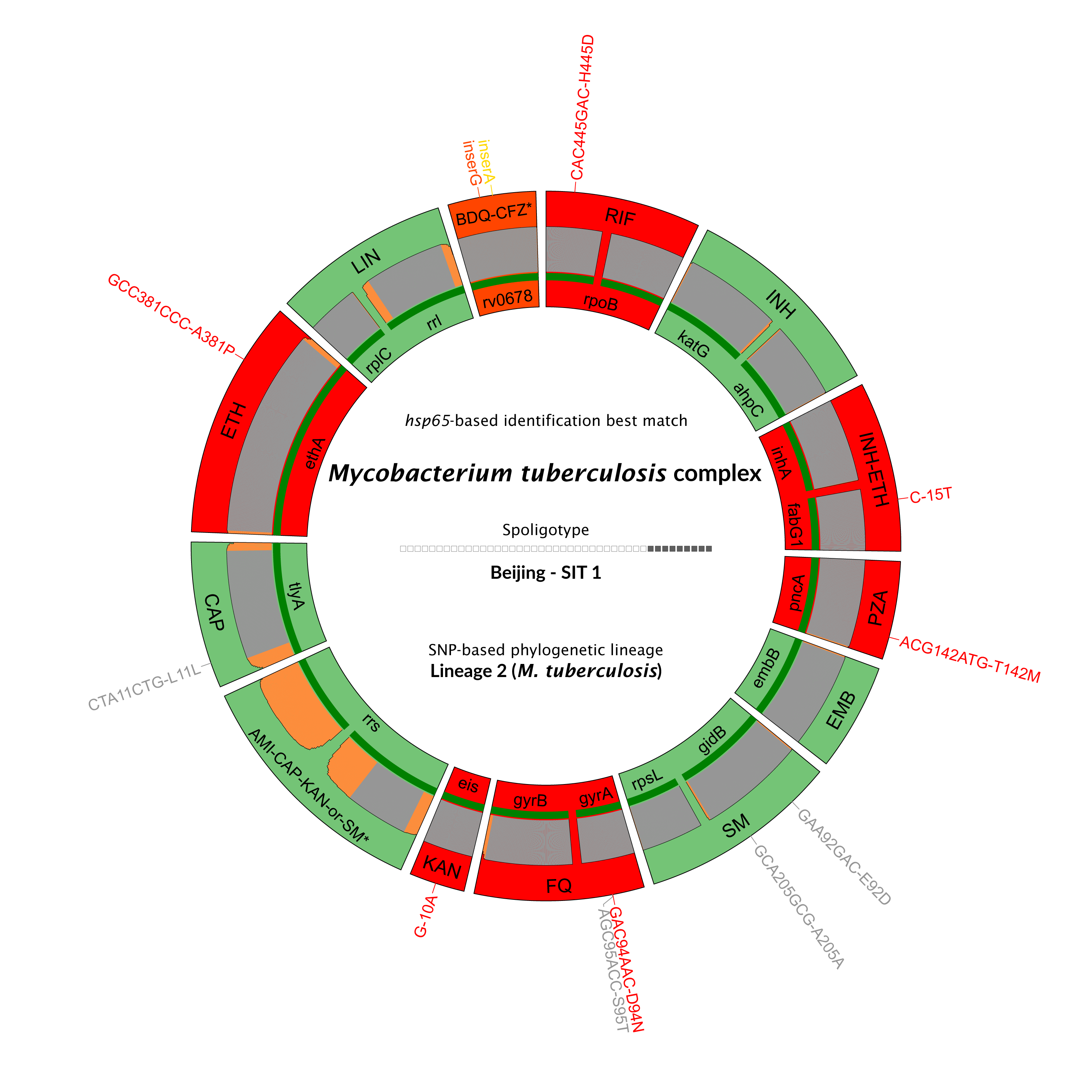
Information on bacterial identification and strain genotype
Gene or genomic region targeted by Deeplex amplification
Drug associated with the gene target
Identified mutations are noticed in red if associated with antimicrobial resistance
Limit of detection and target coverage
Key results at first glance
Get a clear overview of each sample, the Deeplex map visually links antibiotics to genomic regions for rapid and straightforward interpretation.
All detected mutations are mapped for precise identification and extensive prediction of resistance profiles.
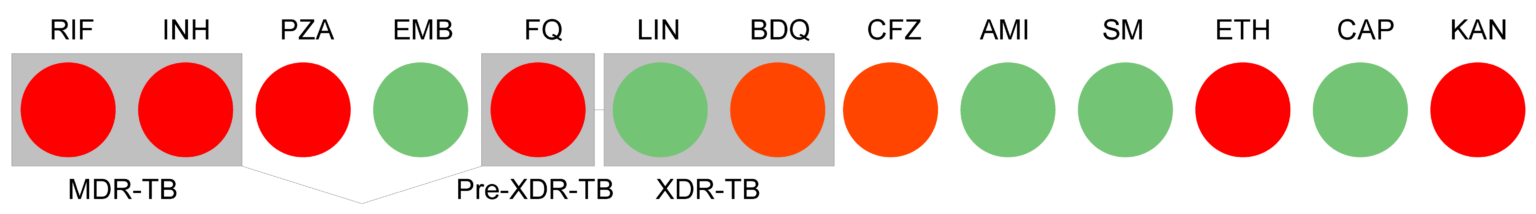
Resistotype: summary of antibiotic resistance prediction
Example of a Deeplex Myc-TB map and its associated resistotype.
Green: No sequence variants identified or variants known not to be linked with antimcrobial resistance.
Red: Identification of one or several variants associated with resistance. The Deeplex map is a registered design.
A streamlined workflow

Sample preparation including DNA extraction from clinical samples or cultured isolates. This step differs depending on the bacteria and the matrix.


Single multiplex PCR for amplification of target gene regions (e.g. for identification, detection of resistance-associated mutations, virulence markers…) within the bacterial genome.

DNA library preparation and deep amplicon sequencing.


Automated analysis of the sequencing data via secure GenoScreen pipelines.


Clear, easy-to-interpret results and reports. No bioinformatics skills required.



